...
The Hierarchical Editing Language for Macromolecules was designed to create a single notation that can encode the structure of complex biomolecules including diverse polymers, non-natural monomers and complex attachment points.
...
HELM was first conceived at Pfizer in the summer of 2008 to support the Pfizer oligonucleotide therapeutic unit and molecules were first registered into the Pfizer corporate database using HELM in December 2008.
How it works
HELM contains multiple levels of information:
Monomers - the atom/bond representation of the building blocks
Simple polymers - a linear sequence of monomers of the same type
Complex polymers - combinations of simple polymers, hydrogen bond information and annotations
...
...
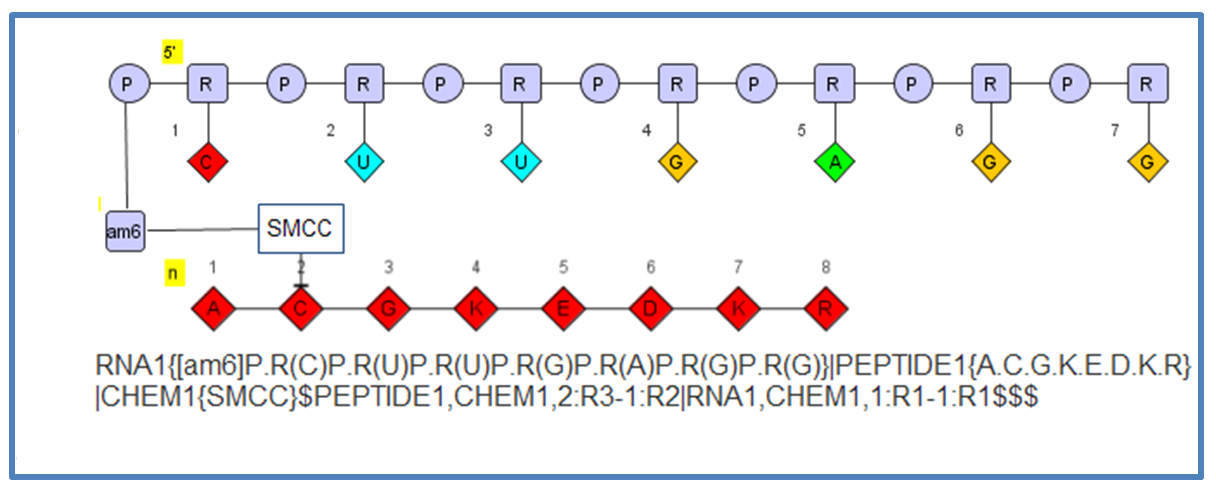
The flexibility allows you to define molecules like this which include a oligonucleotide connected to a small molecule (SMCC) connected to a peptide.
...
There are 2 major releases of HELM.
HELM1 |
|---|
...
HELM 2 | |
|---|---|
Covers different types of macromolecule |
...
: DNA/RNA, |
...
PEPTIDE and |
...
CHEM polymer types |
...
Allows non-natural monomers |
...
: Monomers are defined by the HELM author, so there is no limit on the monomers that can be included in your molecule. Is portable xHELM allows you to ‘bundle’ all the monomer information with your molecule definition into a single package that can be used to transfer information outside your organisation. |
HELM 2
Adds the ability to define
|
Specification
The HELM 2.04 specification is available below.
| View file | ||
|---|---|---|
|
...
There are minor changes in the 2.04 update. Specifically:
Added links to the monomer JSON schema.
Added location of xml schema for xHELM.
Clarified mandatory use of ? for connection points with unknown monomers like *, X and N.
Clarified rules for the use of in-line HELM particularly around connection point definition.
Minor revisions to aid clarity.
Test set
| View file | ||
|---|---|---|
|
...
...
We have developed recommendations for creating and naming a monomer set for peptides.
| View file | ||
|---|---|---|
|
| View file | ||
|---|---|---|
|
An academic overview of HELM and its origins
HELM 1.0 overview
| View file | ||
|---|---|---|
|
...
xHELM overview
| View file | ||
|---|---|---|
|
...
...
HELM 2.0 quick reference diagram
...
...
| View file | ||
|---|---|---|
|
...
Notepad++ highlighting definition file
...
To use this language definition file go to [Language] -> [Define you language...]. Here you have to click on [Import...] and load the xml file. After closing the dialog, you have to restart Notepad++. Now you can assign the language to you current open file via [Language] -> [HELM2].
...