Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
This idea in IP3
https://ip3.pistoiaalliance.org/subdomain/main/end/node/2055
Meeting Minutes
Useful Links
- MLflow
- SAS Model Manager
- Add more here and supply links
Publication References
- Add here and supply links
Project members:
Meeting Minutes and Project Files
Our Publication References
Vladimir A. Makarov, Terry Stouch, Brandon Allgood, Chris D. Willis, Nick Lynch, "Best practices for artificial intelligence in life sciences research", Drug Discovery Today, Volume 26, Issue 5, 2021, Pages 1107-1110, ISSN 1359-6446, https://doi.org/10.1016/j.drudis.2021.01.017. Free pre-print available at https://osf.io/eqm9j/
Abstract: We describe 11 best practices for the successful use of artificial intelligence and machine learning in pharmaceutical and biotechnology research at the data, technology and organizational management levels.
Summary of Our Paper in Drug Discovery Today ("The 11 Commandments")
- DS is not enough: domain knowledge is a must
- Quality data (…and metadata) à quality models
- Long-term data management methodology, e.g. FAIR for life cycle planning for scientific data
- Publish model code, and testing and training data, sufficient for reproduction of research work, along with model results
- Use model management system
- Use ML methods fit for problem class
- Manage executive expectations
- Educate your colleagues – leaders in particular
- AI models + humans-in-the-loop = “AI-in-the-loop” (Chas Nelson invented the term)
- Experiment and fail fast if needed. A bad ML model that is quickly deemed worthless is better than a deceptive model
- Maintain an Innovation Center for moonshot-type technology programs (this COE is an example of one)
Walsh, I., Fishman, D., Garcia-Gasulla, D. et al. "DOME: recommendations for supervised machine learning validation in biology", Nature Methods 18, 1122–1127 (2021). https://doi.org/10.1038/s41592-021-01205-4
Abstract: DOME is a set of community-wide recommendations for reporting supervised machine learning–based analyses applied to biological studies. Broad adoption of these recommendations will help improve machine learning assessment and reproducibility.
Comparison of the Published GMLP Recommendations
How do the Pistoia Alliance recommendations for Good Machine Learning Practices align with those by the leading regulatory authorities?
There is a very considerable overlap between our recommendations (conceived in 2019 and first made public in the April of 2020 as a preprint) and the recommendations by the US FDA and the UK MHRA (published in October 2021). Three out of 10 FDA recommendations follow ours exactly, and six more overlap in content with our recommendations. It is not surprising because both sets of recommendations are based on industry experience and common sense. The remaining difference is due to the difference in the intended audiences: we addressed the ML practitioners whereas the FDA wrote its recommendations for the medical community.
- DS is not enough: domain knowledge is a must (FDA #1 exactly)
- Quality data (…and metadata) à quality models (FDA #3, 4, 5)
- Long-term data management methodology, e.g. FAIR for life cycle planning for scientific data (partly overlaps with FDA #3, 4, 5)
- Publish model code, and testing and training data, sufficient for reproduction of research work, along with model results (partly overlaps with FDA #3, 4, 5, 8, 9)
- Use model management system (not included in the FDA GMLPs, although likely relevant)
- Use ML methods fit for problem class (FDA #6 exactly)
- Manage executive expectations
- Educate your colleagues – leaders in particular
- AI models + humans-in-the-loop = “AI-in-the-loop” (Chas Nelson invented the term; FDA #7 exactly)
- Experiment and fail fast if needed. A bad ML model that is quickly deemed worthless is better than a deceptive model (partly overlaps with FDA #8, 9, 10)
- Maintain an Innovation Center for moonshot-type technology programs
Compare to the US FDA and UK MHRA Good Machine Learning Practice for Medical Device Development: Guiding Principles:
- Multi-Disciplinary Expertise Is Leveraged Throughout the Total Product Life Cycle: In-depth understanding of a model’s intended integration into clinical workflow, and the desired benefits and associated patient risks, can help ensure that ML-enabled medical devices are safe and effective and address clinically meaningful needs over the lifecycle of the device.
- Good Software Engineering and Security Practices Are Implemented: Model design is implemented with attention to the “fundamentals”: good software engineering practices, data quality assurance, data management, and robust cybersecurity practices. These practices include methodical risk management and design process that can appropriately capture and communicate design, implementation, and risk management decisions and rationale, as well as ensure data authenticity and integrity.
- Clinical Study Participants and Data Sets Are Representative of the Intended Patient Population: Data collection protocols should ensure that the relevant characteristics of the intended patient population (for example, in terms of age, gender, sex, race, and ethnicity), use, and measurement inputs are sufficiently represented in a sample of adequate size in the clinical study and training and test datasets, so that results can be reasonably generalized to the population of interest. This is important to manage any bias, promote appropriate and generalizable performance across the intended patient population, assess usability, and identify circumstances where the model may underperform.
- Training Data Sets Are Independent of Test Sets: Training and test datasets are selected and maintained to be appropriately independent of one another. All potential sources of dependence, including patient, data acquisition, and site factors, are considered and addressed to assure independence.
- Selected Reference Datasets Are Based Upon Best Available Methods: Accepted, best available methods for developing a reference dataset (that is, a reference standard) ensure that clinically relevant and well characterized data are collected and the limitations of the reference are understood. If available, accepted reference datasets in model development and testing that promote and demonstrate model robustness and generalizability across the intended patient population are used.
- Model Design Is Tailored to the Available Data and Reflects the Intended Use of the Device: Model design is suited to the available data and supports the active mitigation of known risks, like overfitting, performance degradation, and security risks. The clinical benefits and risks related to the product are well understood, used to derive clinically meaningful performance goals for testing, and support that the product can safely and effectively achieve its intended use. Considerations include the impact of both global and local performance and uncertainty/variability in the device inputs, outputs, intended patient populations, and clinical use conditions.
- Focus Is Placed on the Performance of the Human-AI Team: Where the model has a “human in the loop,” human factors considerations and the human interpretability of the model outputs are addressed with emphasis on the performance of the Human-AI team, rather than just the performance of the model in isolation.
- Testing Demonstrates Device Performance during Clinically Relevant Conditions: Statistically sound test plans are developed and executed to generate clinically relevant device performance information independently of the training data set. Considerations include the intended patient population, important subgroups, clinical environment and use by the Human-AI team, measurement inputs, and potential confounding factors.
- Users Are Provided Clear, Essential Information: Users are provided ready access to clear, contextually relevant information that is appropriate for the intended audience (such as health care providers or patients) including: the product’s intended use and indications for use, performance of the model for appropriate subgroups, characteristics of the data used to train and test the model, acceptable inputs, known limitations, user interface interpretation, and clinical workflow integration of the model. Users are also made aware of device modifications and updates from real-world performance monitoring, the basis for decision-making when available, and a means to communicate product concerns to the developer.
- Deployed Models Are Monitored for Performance and Re-training Risks are Managed: Deployed models have the capability to be monitored in “real world” use with a focus on maintained or improved safety and performance. Additionally, when models are periodically or continually trained after deployment, there are appropriate controls in place to manage risks of overfitting, unintended bias, or degradation of the model (for example, dataset drift) that may impact the safety and performance of the model as it is used by the Human-AI team.
Business Analysis
Our business analysis is based on enumeration of business personas and the challenges that these personas may face in their work with AI/ML technologies. The proposed Good Machine Learning Practices (GMLPs) answer these challenges.
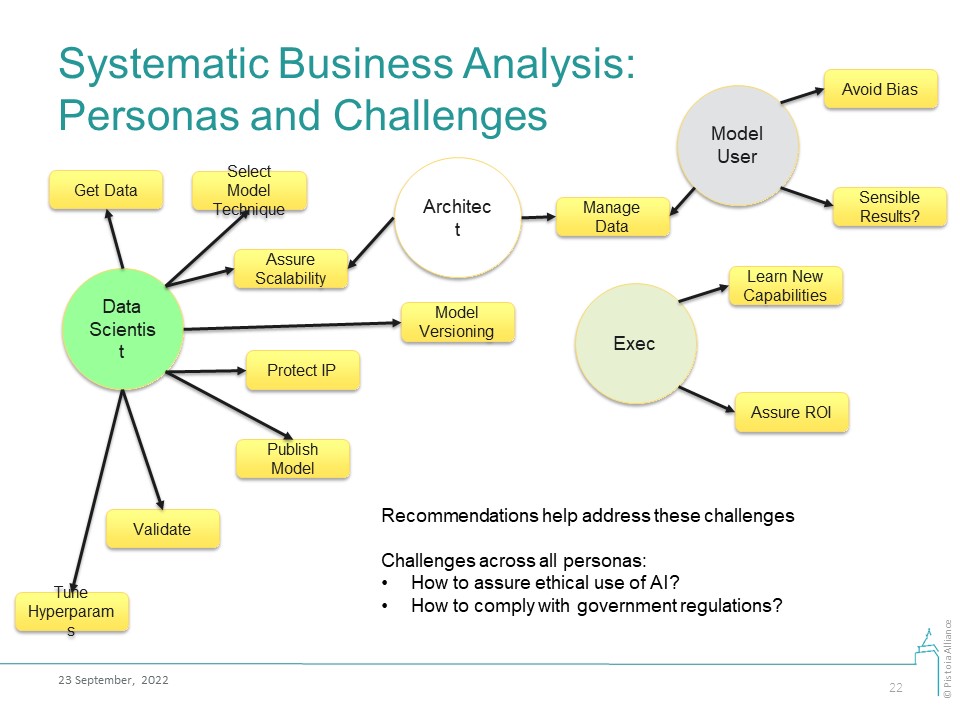 Image Added
Image Added
Persona | Challenges | GMLPs that address these challenges | Author(s) | Reviewer(s) |
Data Scientist | DS1: How do I pick a suitable data modeling technique? | 1. Make recommendations for methods suitable for specific problem classes; or for auto-ML systems 2. DS should learn the application domain and/or work with domain experts | ||
DS2: How do I store and version a model? | 1. Recommend model versioning systems 2. Refer to best practices in ML Model Registration, Deployment, Monitoring 3. Best practices and regulatory standards in requirements management and model provenance | Christophe Chabbert | ||
| Samuel Ramirez | |||
1. Refer to the DOME recommendations 2. Quickly fail models that are bad. A deceptive model is worse than a bad one | Fotis Psomopoulos | |||
Elina Koletou | ||||
1. Recommend best practices from the FAIR Toolkit 2. Evaluate data for “fit for purpose”, in particular, for the metadata quality 3. Refer to best practices in Exploration, Cleaning, Normalizing, Feature Engineering, Scaling | Natalja Kurbatova, Christophe Chabbert, Berenice Wulbrecht | |||
Christophe Chabbert | ||||
DS8: How do I publish AI models? | Publish model code, and testing and training data, sufficient for reproduction of research work, along with model results. Make recommendations for an appropriate minimal information model | Fotis Psomopoulos | ||
DS9: How to protect IP? | ||||
DS10: How should the applicability domain of the model be expressed? | Use Applicability Domain Analysis | Samuel Ramirez | ||
Model User | MS1: How should I manage the data? Includes data protection, versioning, labeling | 1. Recommend best practices from the FAIR Toolkit 2. Evaluate data for “fit for purpose”, in particular, for the metadata quality 3. Refer to best practices in Exploration, Cleaning, Normalizing, Feature Engineering, Scaling | Natalja Kurbatova, Christophe Chabbert, Berenice Wulbrecht | |
MS2: How do I avoid bias? | ||||
MS3: How do I make sure the model produces sensible answers? | 1. Set-up a “human-in-the-loop” system. Recommend tools for this, if they exist 2. Set-up business feedback mechanism for flagging model results that do not align with expectations | Chas Nelson | ||
Architect | Christophe Chabbert | |||
See above for Model User | Christophe Chabbert | |||
1. Refer to best practices in DevOps 2. Automated model packaging for ease of production delivery | Elina Koletou | |||
Elina Koletou | ||||
Executive | E1: How do I learn about the costs and benefits of AI/ML technologies and the limits of possible? | Make recommendations for conferences, training materials, education products, review papers, and books. These must be updated on a frequent cadence | ||
E2: How do I value and justify investments in AI? | 1. Make recommendations for technology valuation methodologies 2. Provide business questions, goals, KPIs 3. Continuously review model performance against the business needs (goals, KPIs) | Prashant Natarajan | ||
E3: How do I enable technology adoption? | 1. Bring expertise together to assess compatibility of new technology with business architecture 2. Ensure sufficient knowledge transfer so users are comfortable with adoption and use the new approach 3. Have a sustainability plan so resources are available when needed | Prashant Natarajan |
Common challenges across ALL personas:
- How do I develop and use AI/ML models in an ethical manner (privacy protection, ethical use, full disclosure, etc)?
- How do I develop and use AI/ML models in compliance with the government regulations?
Common responsibility across multiple personas:
- Model improvement is a shared responsibility between the Model User, Architect, and Data Scientist personas.
Glossary notes: In the context of this document these terms have these meanings:
- Performance - metrics used to evaluate model quality in terms of sensitivity and specificity, AOC, or similar metrics, or execution speed.
- Validation - test to confirm performance of the model (as defined above); but should not be confused with “validation” in regulated systems.
Project members:
Fotis | Psomopoulos | CERTH |
Brandon | Allgood | Valo Health |
Christophe | Chabbert | Roche |
Adrian | Schreyer | Exscientia |
Elina | Koletou | Roche |
Frederik | van der Broek | Elsevier |
David | Wöhlert | Elsevier |
John | Overington | Exscientia |
Loganathan | Kumarasamy | Zifo R&D |
Neal | Dunkinson | Scibite |
Simon | Thornber | GSK |
Irene | Pak | BMS |
Berenice | Wullbrecht | Ontoforce |
Prashant | Natarajan | H2O.ai |
Valerie | Morel | Ontoforce |
Yvonna | Li | Roche |
Silvio | Tosatto | Unipd.it |
Natalja | Kurbatova | Zifo R&D |
Mufis | Thalath | Zifo R&D |
Niels | Van Beuningen | Vivenics |
Ton | Van Daelen | BIOVIA (3ds) |
Lucille | Valentine | Gliff.ai |
Adrian | Fowkes | Lhasa |
Chas | Nelson | Gliff.ai |
Paolo | Simeone | Ontoforce |
| Christoph | Berns | Bayer |
| Mark | Earll | Syngenta |
Vladimir | Makarov | Pistoia Alliance |
Useful Links
EU Artificial Intelligence Act: https://www.consilium.europa.eu/en/press/press-releases/2022/12/06/artificial-intelligence-act-council-calls-for-promoting-safe-ai-that-respects-fundamental-rights/
US FDA SaMD Action Plan (September 2022): https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device; also see announcement at https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device or download PDF at https://www.fda.gov/media/145022/download
- UK MHRA Software as a Medical Device: https://www.gov.uk/government/publications/software-and-ai-as-a-medical-device-change-programme/software-and-ai-as-a-medical-device-change-programme-roadmap
Related topic at the Pistoia Alliance, a FAIR Guide for Clinical Data: FAIR4Clin
- The proof of the pudding: in praise of a culture of real-world validation for medical artificial intelligence
- The need to separate the wheat from the chaff in medical informatics: Introducing a comprehensive checklist for the (self)-assessment of medical AI studies
- MLflow
- SAS Model Manager
- Promoting the Use of Trustworthy Artificial Intelligence in the Federal Government. Executive Order 13960 of December 3, 2020
- Artificial Intelligence and Machine Learning (AI/ML) for Drug Development. FDA paper
- Good Machine Learning Practice for Medical Device Development: Guiding Principles. US FDA GMLP guide. https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles
- Products of the AFDO GMLP tem:
- https://www.healthcareproducts.org/ai/working-teams/gmlp/
- https://www.healthcareproducts.org/bias-in-healthcare-artificial-intelligence-examined-in-whitepaper/
- https://www.healthcareproducts.org/ai/working-teams/gmlp/data-quality-whitepaper/
- https://www.healthcareproducts.org/wp-content/uploads/2022/04/AI_Whitepaper_BuildingExplainability_final3.pdf