Peptide and Nucleotide Ambiguous Monomers
There are four ambiguous monomers in HELM
* represents 0-n unknown monomers
X, N represents a single unknown monomer. X is used in peptide polymers and N in RNA polymers
_ represents a missing monomer
General principles
Ambiguous monomers exist within a polymer type. Therefore, you can have a PEPTIDE * monomer which indicates that you know it is a section of peptide, but you don’t know anything else.
If you don’t even know that much, then you can opt for CHEM{*} or BLOB. Advice on using these are in a separate ‘best practices’ guide.
Peptide ambiguous monomers
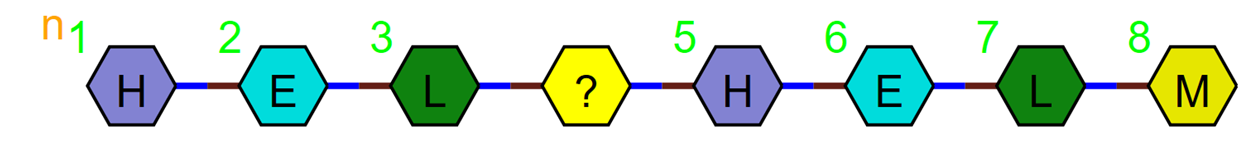
If nothing is known about the ‘?’ then you should use *. PEPTIDE1{H.E.L.*.H.E.L.M}$$$$V2.0
This means we don’t know how long the unknown section is, just that it is made up of peptide monomers.
If we know there is only one monomer, you can use X as in PEPTIDE1{H.E.L.X.H.E.L.M}$$$$V2.0
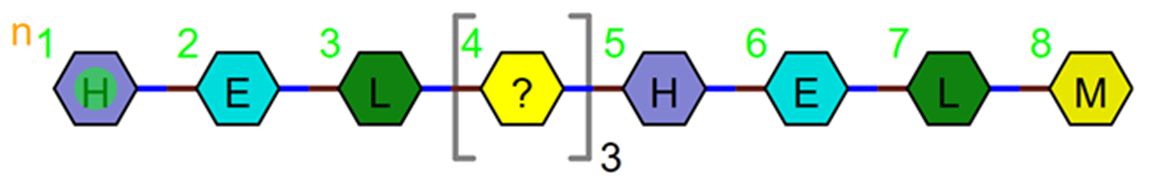
But if there is more than one unknown monomer, and you know how many, use brackets.
The repeat length can be exact e.g. PEPTIDE1{H.E.L.X'3'.H.E.L.M}$$$$V2.0
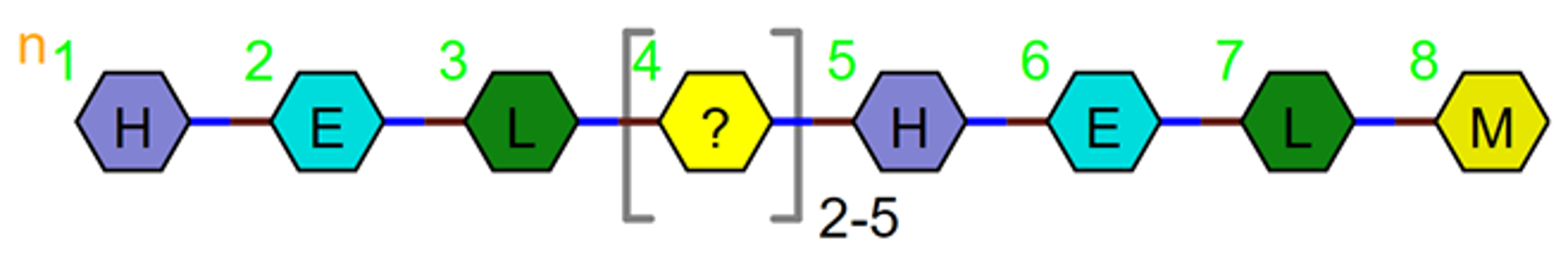
or a range e.g. PEPTIDE1{H.E.L.X'2-5'.H.E.L.M}$$$$V2.0
Missing peptide monomers
The character ‘_’ can be used to show a missing monomer. However, this makes no sense if it is used in a standard HELM string.
PEPTIDE1{H.E.L._.H.E.L.M}$$$$V2.0
is identical to
PEPTIDE1{H.E.L.H.E.L.M}$$$$V2.0,
So, there is no point.
Missing peptide monomers are useful when you are specifying that a monomer may or may not have been included in a polymer. Example: A trailing Lysine might be present at the C-terminus of an antibody or not.
PEPTIDE1{H.E.L.M.H.E.L.M.(K,_)}$$$$V2.0
Nucleotides
Nucleotides are more complex than peptides since the repeating unit is not a single monomer, but three connected together.
*
As for peptides, a ‘*’ monomer represents one or more monomers, so there is no way of knowing whether this includes multiple triad, or a single sugar.
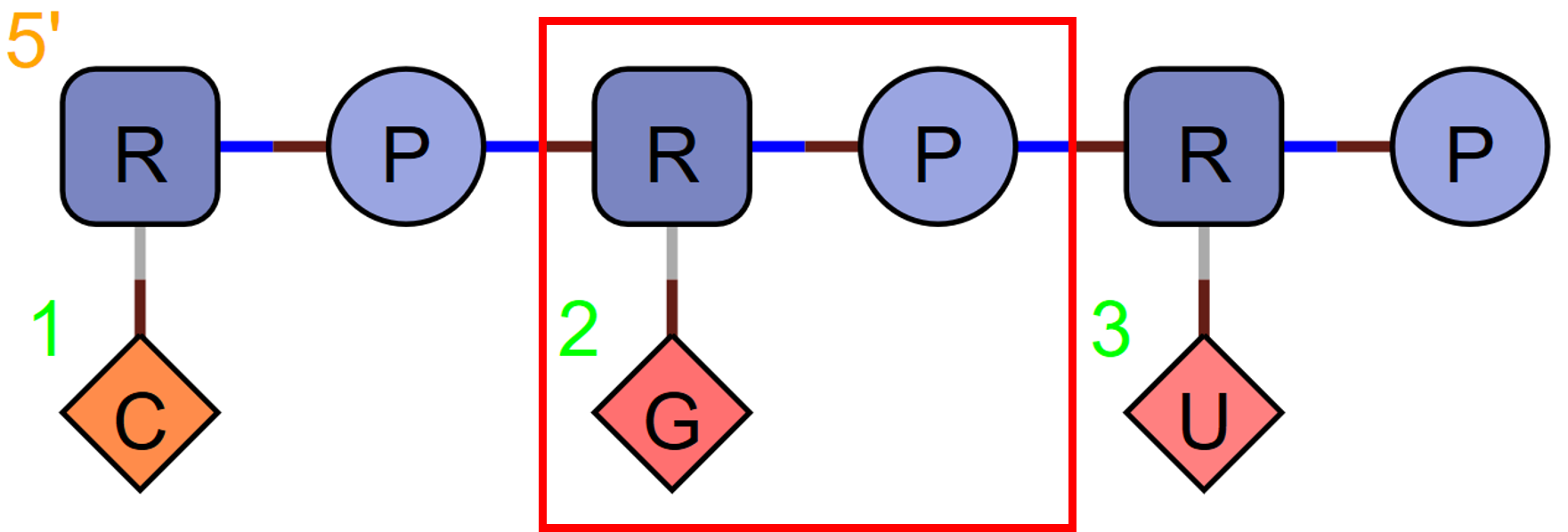
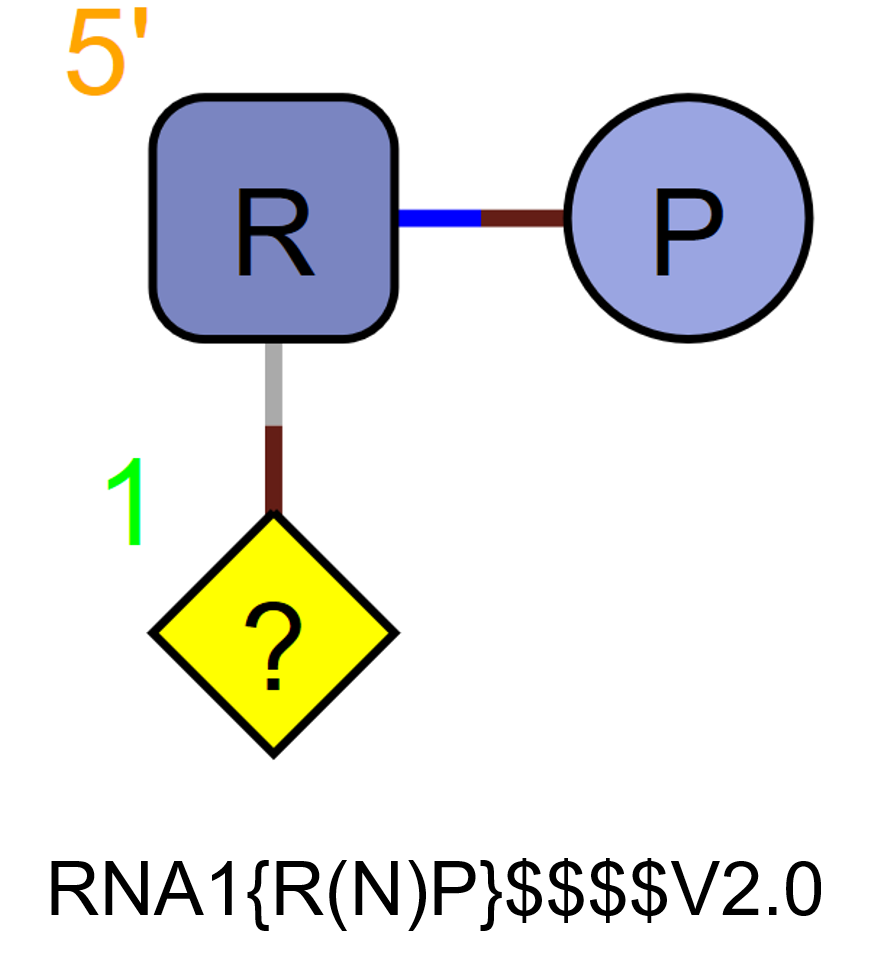
N
N represents a single nucleotide base. It is used alongside the sugar and phosphate and does not replace them. It does not represent a triad.
RNA1{R(N)P}$$$$V2.0
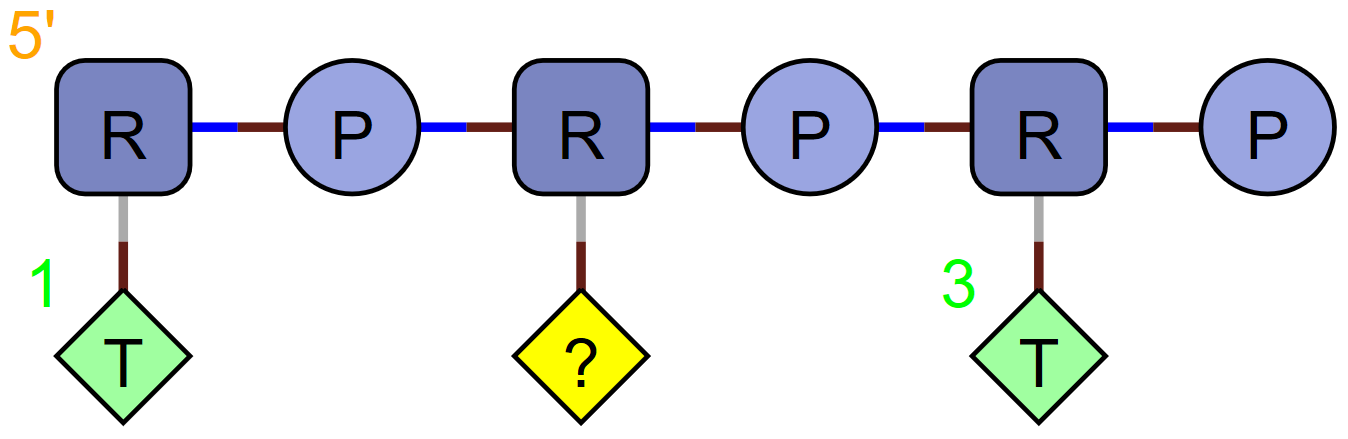
Missing Monomer ‘_’
Missing monomers represent a missing base, so the backbone must still be specified, but the base is missing.
RNA1{R(T)P.R(_)P.R(T)P}$$$$V2.0
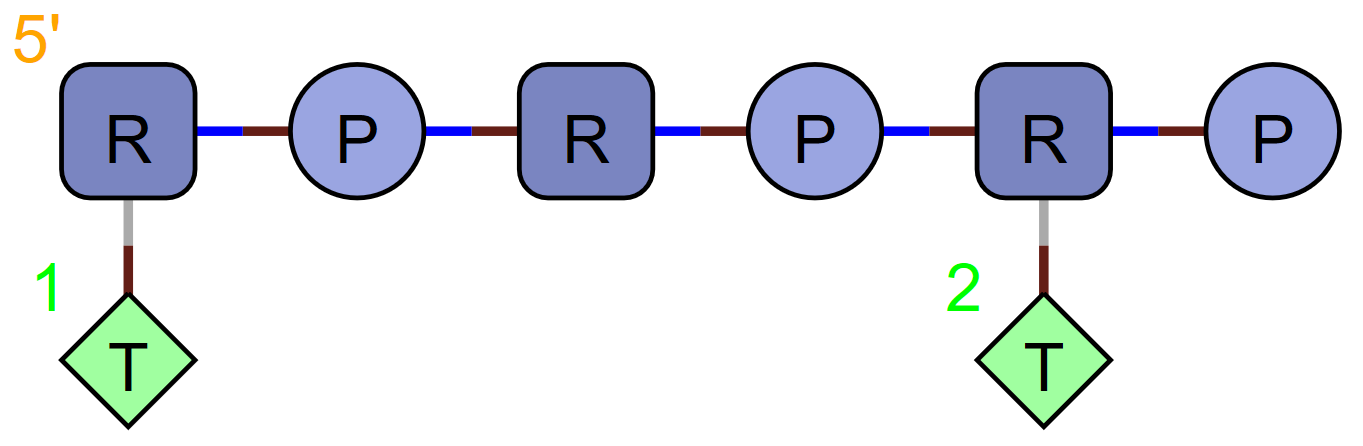
As with peptides, there is little point since you can also represent this molecule using standard HELM like this:
RNA1{R(T)P.RP.R(T)P}$$$$V2.0
A missing monomer symbol is more useful when you are not sure if a base is present or not. You can list the missing base with other options.
RNA1{R(T)P.R(_,A)P.R(T)P}$$$$V2.0
The ‘_’ symbol does not represent a missing triad.