HELM Parsers and Writers
HELM Parsers and Writers
Many organisations have macromolecule information in a variety of formats. Converting HELM to an atom/bond molecular representation is a relatively straightforward process since the monomers and attachment points between monomers are defined, so assembly is straightforward.
Converting an atom/bond representation to HELM is more complex, since you have to decide what the monomers are.
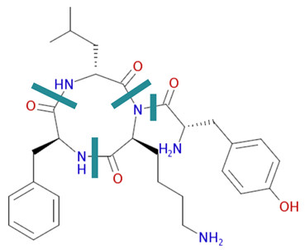
Identifying monomers is not always straightforward. There are decisions about where the backbone actually is which is not always straightforward. See this cyclic peptide as an example:
And there are decisions about capping groups; whether groups such as N-methyl lysine are single monomers or lysine is one monomer and the methyl group a second one.
Free to use toolsets
The HELM toolkit contains a basic fragmenter, and the EBI has completed some work in this area, details of which would be available from them on request.
RDKit
RDKit (open source cheminformatics software released under the BSD license) now includes HELM readers and writers for peptides courtesy of Roger Sayle of NextMove Software. This adds HELM as a supported format to the existing options of SMILES/SMARTS, SDF, TDT, SLN, Corina mol2 and PDB.
A brief overview can be found in some slides presented at the RDKit 2015 user group meeting – see slides 18-22.
https://github.com/rdkit/UGM_2015/tree/master/Presentations
RDKit Github project can be found at:
https://github.com/rdkit/rdkit
The specific code is at:
rdkit/Code/GraphMol/FileParsers/SequenceParsers.cpp
rdkit/Code/GraphMol/FileParsers/SequenceWriters.cpp
Commercial converters
Below is a list of the commercial conversion tools we are aware of. If you know of or indeed sell a commercial HELM conversion tool, please let us know at info@openhelm.org and we will add it to this list.
NextMove Software's Sugar & Splice.
Sugar and Splice supports peptides along with DNA/RNA, inline HELM and XHELM [and IUPAC name generation, and depiction, and peptide name generation, and variable attachment points, and oligosaccharides etc.]. Further details can be found at https://www.nextmovesoftware.com/sugarnsplice.html
ChemAxon Biomolecule Toolkit
ChemAxon provides various conversion utilities from and to HELM within their Biomolecule Toolkit for peptide- and nucleic acid-derived molecules . (https://www.chemaxon.com/products/biomolecule-toolkit/)
Supported file formats for exporting include XHELM, FASTA, plain sequences and MDL/Mol files*. Sequences from HELM are generated using the Natural Analogue information of HELM monomers. Mol files will be exported in v3000 format containing S-group abbreviations for each monomer.
Supported file formats for importing include FASTA, plain sequences and MDL/Mol files*. FASTA headers are maintained in the HELM annotation section. Import of Mol files is currently limited to molecules, which are fully represented by a given monomer dictionary. This dictionary may be a fully custom/proprietary one.
Until mid 2016 import of PDB files is expected to become available.
*Other chemical file formats can be converted to MDL Mol-file representation using ChemAxon's Molconvert.
Biovia Pipeline Pilot
Biovia's Pipeline Pilot contains HELM reader and writer components in its chemistry collection. It does not fragment atom/bond representations but can convert sequence-based notations. There is HELM support in some other Biovia tools - see the PDF below for details.
Perkin-Elmer's ChemDraw
Version 17 of ChemDraw allows you to draw HELM molecules directly using the monomer.org monomer set. A video is available to show you how.
.